L'Eric Topol ens ofereix un resum d'alta qualitat per entendre el moment que viu la ciència mitjançant la intel·ligència artificial.
Impressionant. Aquesta és la llista de grans models de llenguatge en ciències de la vida (LLLMs):
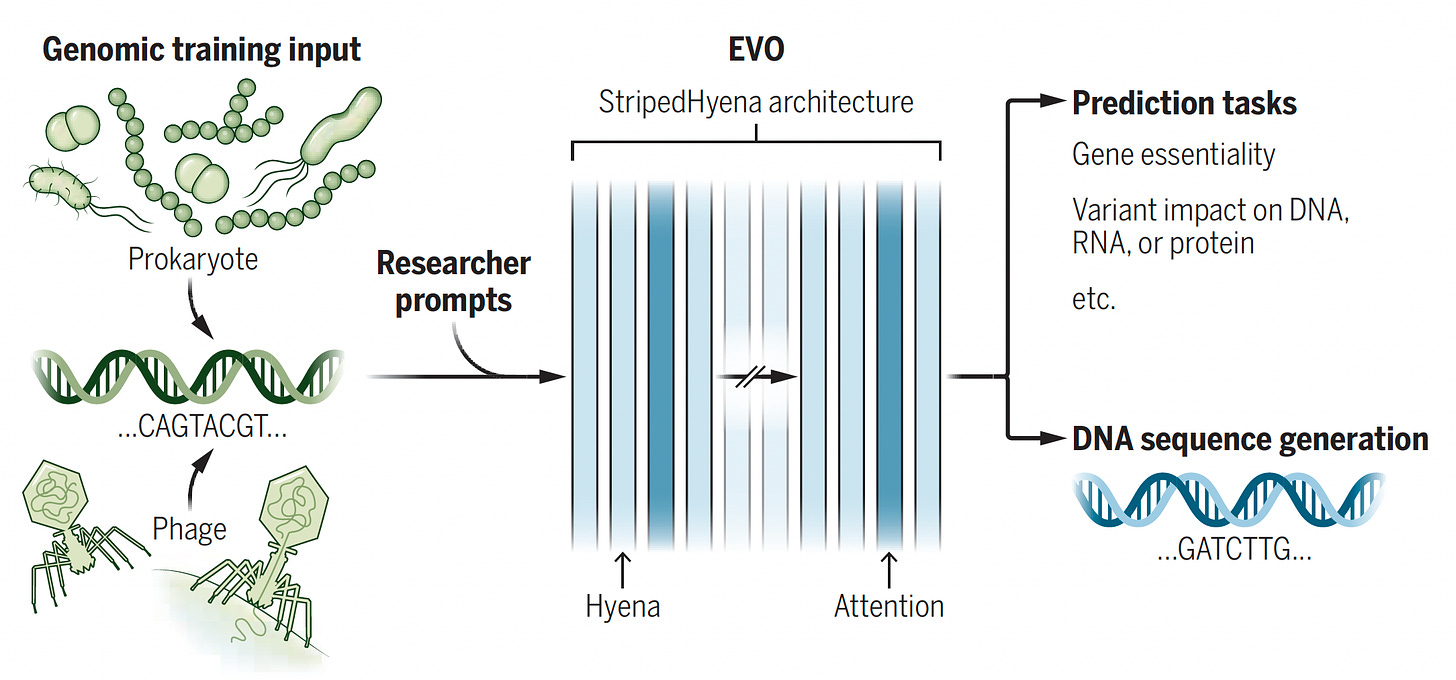
Evo. This model was trained with 2.7 million evolutionary diverse organisms (prokaryotes—without a nucleus, and bacteriophages) representing 300 billion nucleotides to serve as a foundation model (with 7 billion parameters) for DNA language, predicting function of DNA, essentiality of a gene, impact of variants, and DNA sequence or function, and CRISPR-Cas prediction. It’s multimodal, cutting across protein-RNA and protein-DNA design.
Figure below from accompanying perspective by Christina Theodoris.
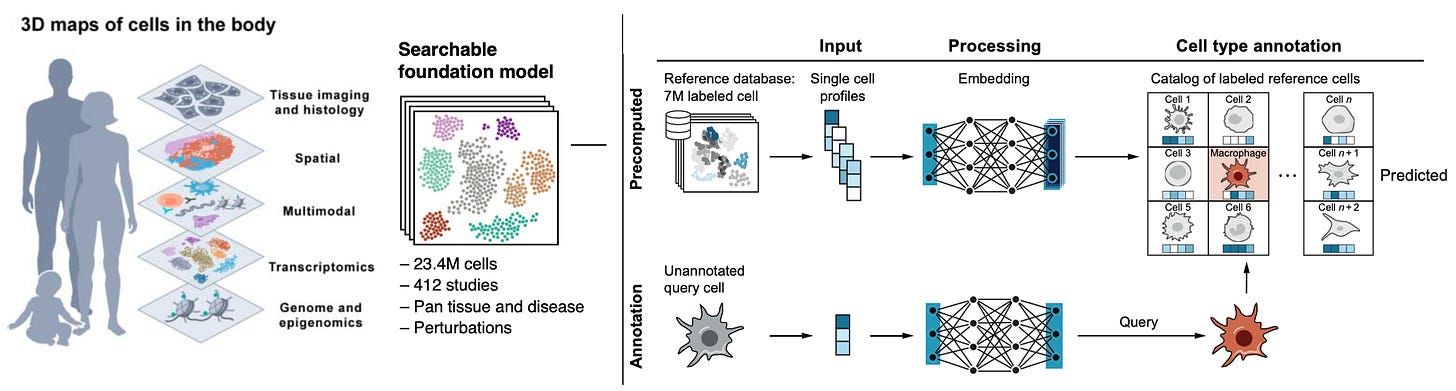
Human Cell Atlas A collection of publications from this herculean effort involving 3,000 scientists, 1,700 institutions, and 100 countries, mapping 62 million cells (on the way to 1 billion), with 20 new papers that can be found here. We have about 37 trillion cells in our body and until fairly recently it was thought there were about 200 cell types. That was way off—-now we know there are over 5,000.
One of the foundation models built is Single-Cell (SC) SCimilarity, which acts as a nearest neighbor analysis for identifying a cell type, and includes perturbation markers for cells (Figure below). Other foundation models used in this initiative are scGPT, GeneFormeR, SC Foundation, and universal cell embedding. Collectively, this effort has been called th “Periodic Table of Cells” or a Wikipedia for cells and is fully open-source. Among so many new publications, a couple of notable outputs from the blitz of new reports include the finding of cancer-like (aneuploid) changes in 3% of normal breast tissue, representing clones of rare cells and metaplasia of gut tissue in people with inflammatory bowel disease.
BOLTZ-1 This is a fully open-source model akin to AlphaFold 3, with similar state-of-the-art performance, for democratizing protein-molecular interactions as summarized above (for AlphaFold 3). Unlike AlphaFold 3 which is only available to the research community, this foundation model is open to all. It also has some tweaks incorporated beyond AlphaFold 3, as noted in the preprint.
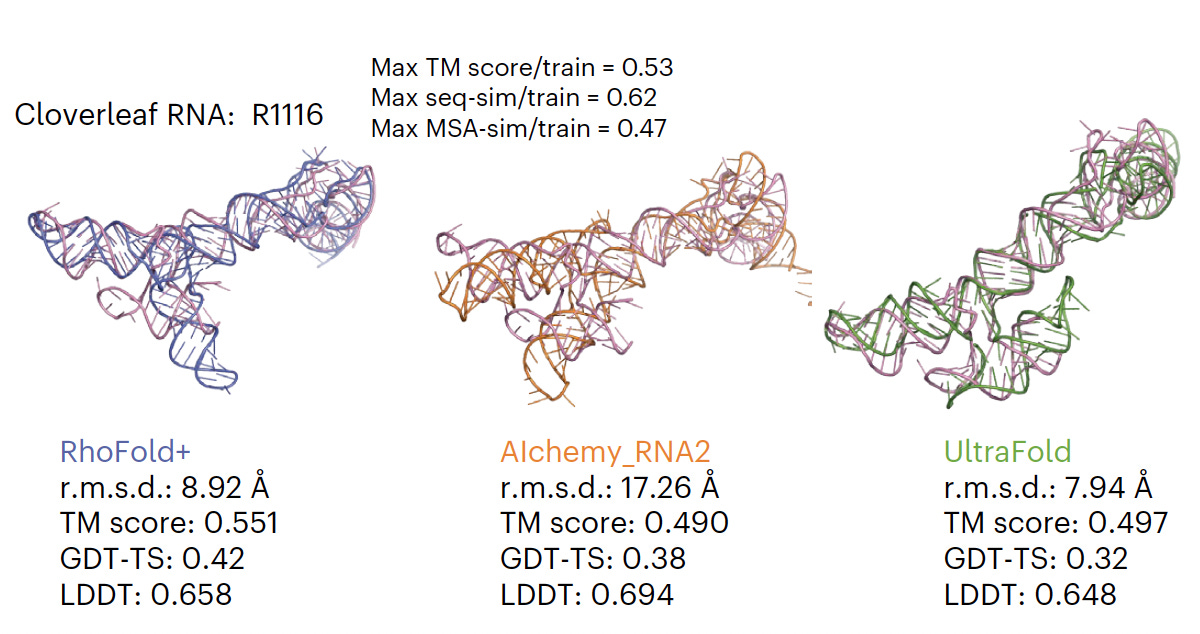
RhoFold For accurate 3D RNA structure prediction, pre-trained on almost 24 million RNA sequences, superior to all existing models (as shown below for one example).
EVOLVEPro A large language protein model combined with a regression model for genome editing, antibody binding and many more applications for evolving proteins, all representing a jump forward for the field of A.I. guided protein engineering.
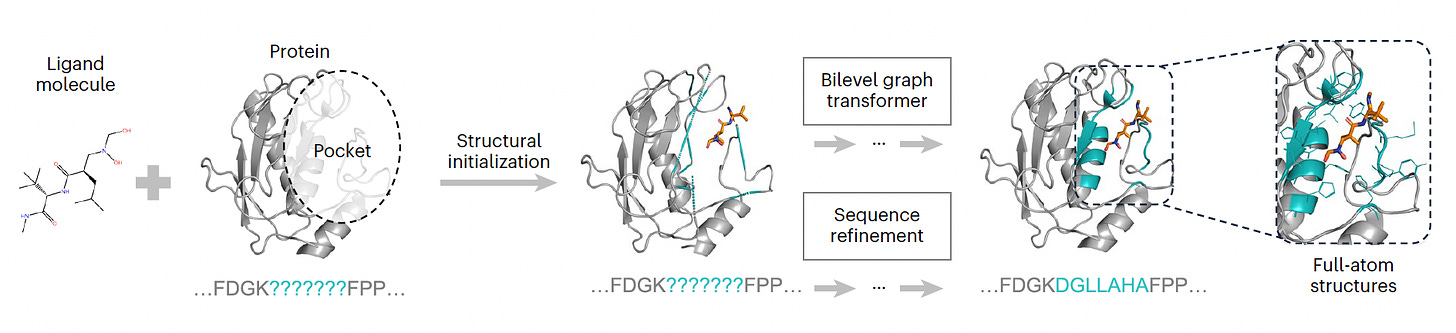
PocketGen A model dedicated to defining the atomic structure of protein regions for their ligand interactions, surpassing all previous models for this purpose.
MassiveFold A version of AlphaFold that does predictions in parallel, enabling a marked reduction of computing time from several months to hours
RhoDesign From the same team that produced RhoFold, but this model is for efficient design of RNA aptamers that can be used for diagnostics or as a drug therapy.
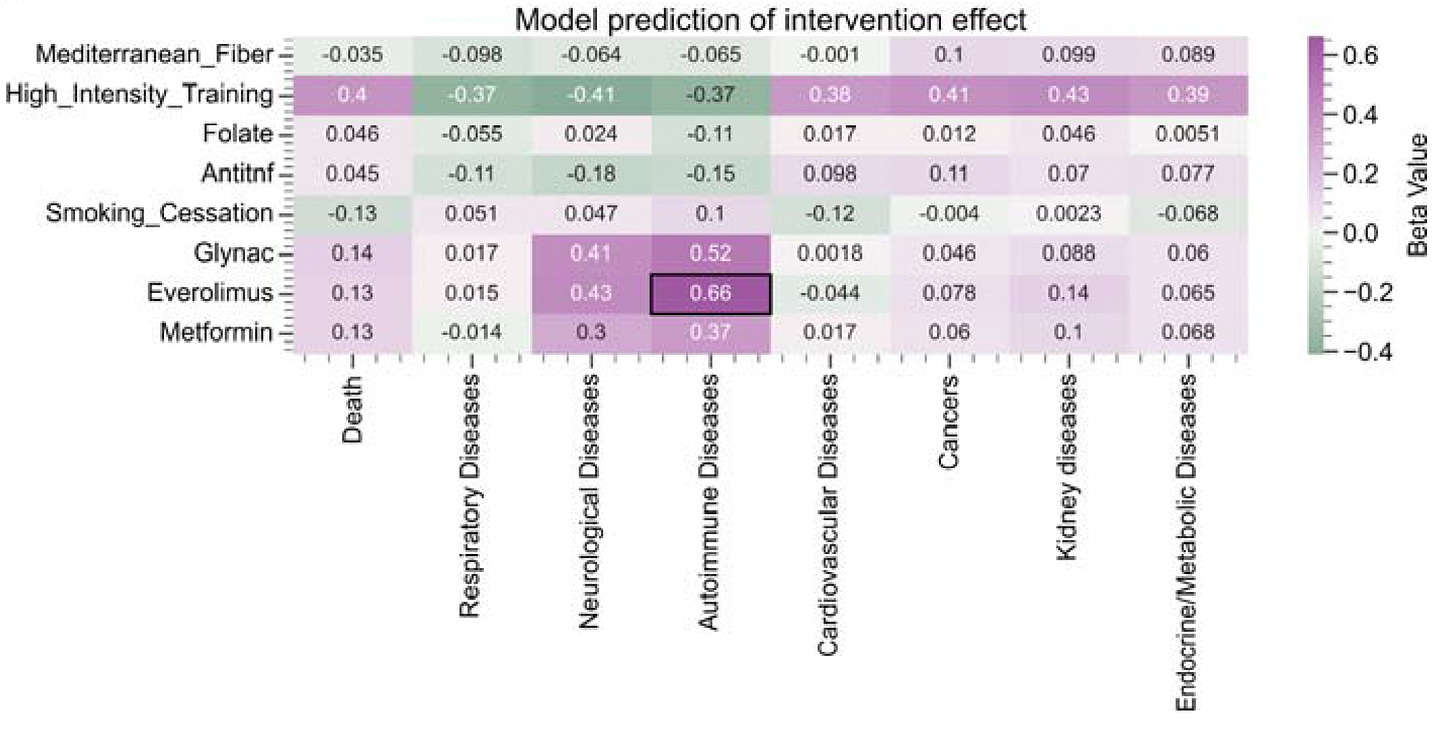
MethylGPT Built upon scGPT architecture, trained on over 225,000 samples, it captures and can reconstruct almost 50,000 relevant methylation CpG sites which help in predicting diseases and gauging the impact of interventions (see graphic below).
CpGPT Trained on more than 100,000 samples, it is the optimal model to date fo predicting biological (epigenetic) age, imputing missing data, and understanding biology of methylation patterns.
PIONEER A deep learning pipeline dedicated to the protein-protein interactome, identifying almost 600 protein-protein interactions (PPIs) from 11,000 exome sequencing across 33 types of cancer, leading to the capability of prediction which PPIs are associated with survival. (This was published 24 October, the only one not in November on the list!)