If epigenomics is crucial to discard the genetic predestination paradigm, now we can add a new 'omics to the paradigm: epitranscriptomics. Last February, Nature published interesting news related to recent scientific developments:
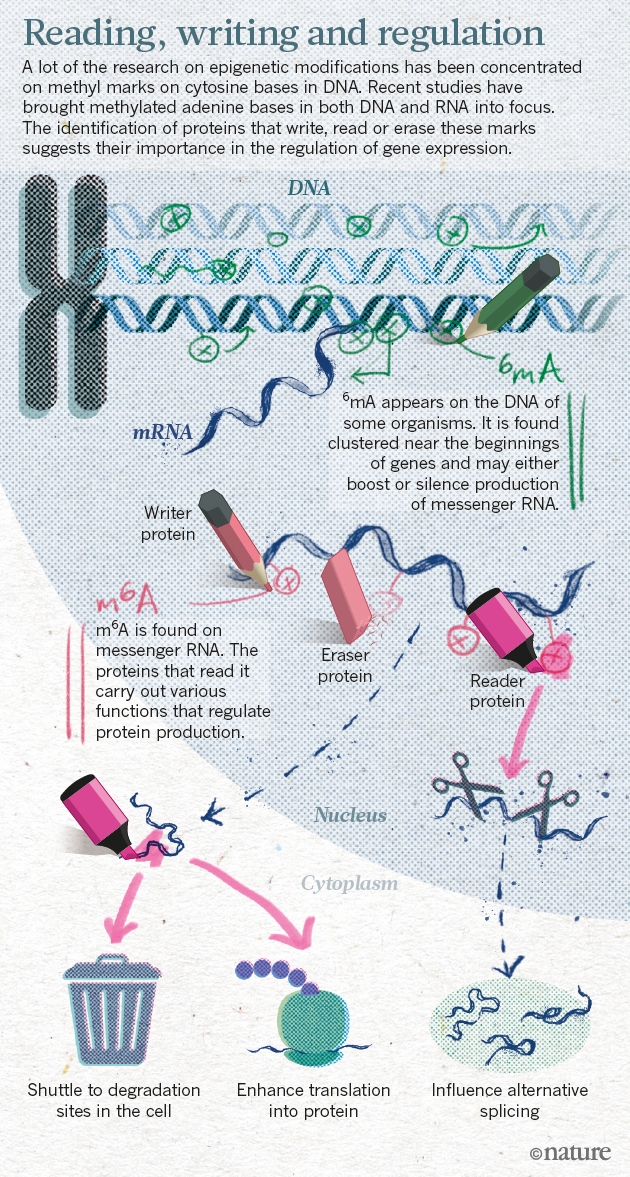
The epigenome helps to explain how cells with identical DNA can develop into the multitude of specialized types that make up different tissues. The marks help cells in the heart, for example, maintain their identity and not turn into neurons or fat cells. Misplaced epigenetic marks are often found in cancerous cells.Chuan He and Tao Pan are two researchers that have been working on new ways of controlling gene expression
He and others have shown that a methyl group attached to adenine, one of the four bases in RNA, has crucial roles in cell differentiation, and may contribute to cancer, obesity and more. In 2015, He’s lab and two other teams uncovered the same chemical mark on adenine bases in DNA (methyl marks had previously been found only on cytosine), suggesting that the epigenome may be even richer than previously imagined.
The team had shown for the first time that RNA methylation was reversible, just like the marks found on DNA and histones.Methylated adenine bases are the focus of research on gene expression.