Epigenetic protein families: a new frontier for drug discovery
Si ja sabem que al costat de la genòmica hi tenim la proteòmica i la metabolòmica, i que per sobre encara hi tenim l'epigenètica (que literalment vol dir més enllà de la genètica), ara a
Nature ens expliquen les diferent famílies i el que representen per al futur de la recerca en nous medicaments.
L'article de revisió és d'aquells que em guardaré perquè si fins ara s'explicava la importància de l'epigènetica i com el paradigma genòmic de la predestinació brandava com un saltamartí, calia posar ordre a les idees. Però també perquè explica amb tot detall com:
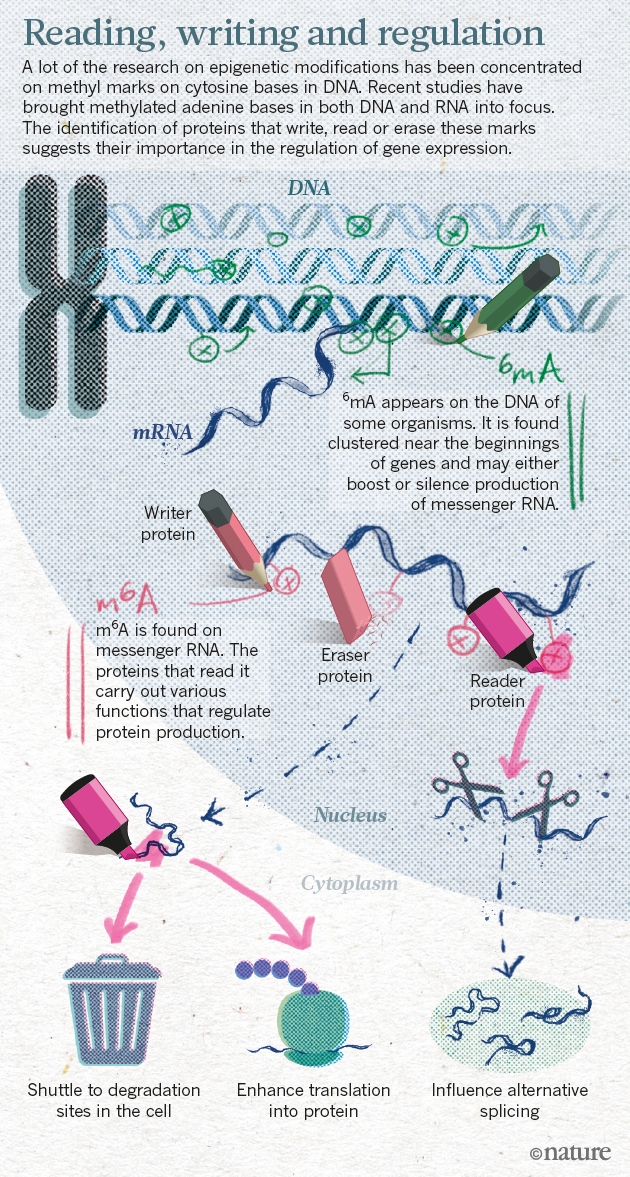
Epigenetic regulation of gene expression is a dynamic and reversible process that establishes normal cellular phenotypes but also contributes to human diseases. At the molecular level, epigenetic regulation involves hierarchical covalent modification of DNA and the proteins that package DNA, such as histones. Here, we review the key protein families that mediate epigenetic signalling through the acetylation and methylation of histones, including histone deacetylases, protein methyltransferases, lysine demethylases, bromodomain-containing proteins and proteins that bind to methylated histones. These protein families are emerging as druggable classes of enzymes and druggable classes of protein–protein interaction domains.
L'explicació inicial m'ha semblat un resum útil:
Although all cells in an organism inherit the same genetic material, the ability of cells to maintain the unique physical characteristics and biological functions of specific tissues and organs is due to heritable differences in the packaging of DNA and chromatin. These differences dictate distinct cellular gene expression programmes but do not involve changes in the underlying DNA sequence of the organism. Thus, epigenetics (which literally means ‘above genetics’) underpins the fundamental basis of human physiology. Importantly, the epigenetic state of a cell is malleable; it evolves in an ordered manner during the cellular differentiation and development of an organism, and epigenetic changes are responsible for cellular plasticity that enables cellular reprogramming and response to the environment. Because epigenetic mechanisms are responsible for the integration of environmental cues at the cellular level, they have an important role in diseases related to diet, lifestyle, early life experience and environmental exposure to toxins1. Thus, epigenetics is of therapeutic relevance in multiple diseases such as cancer, inflammation, metabolic disease and neuropsychiatric disorders, as well as in regenerative medicine
Així doncs, ens trobem davant un horitzó de noves descobertes que es va configurant i que explica en bona part perquè s'ha tardat més d'una dècada en traslladar el projecte genoma humà cap a aplicacions terapèutiques àmplies. Però també s'obre un nou interrogant sobre a seguretat dels modificadors epigenètics dels medicaments. La forma com caldrà avaluar-ho suposarà més exigència al regulador i una necessitat de transparència de la caixa negra encara més gran. Seguirem atents, perquè per ara ja se li ha girat feina amb els inhibidors HDAC, per al limfoma cutani de cèl.lules T, el primer d'aquests medicaments.
PS. I si voleu una perspectiva diferent, consulteu
aquest article.
PS. Millor no saber-ho. Ja ho vaig explicar fa temps i ara ho trobareu al
WSJ. Cal conèixer la teva predisposició a l'Alzheimer mitjançant un test genètic? de què et servirà. Podeu llegir
aquí una història real que em confirma el que deia.
PS. I si l'altre dia teníem una
galleda d'aigua freda, ara en tenim una altra de calenta.
23andMe acaba d'obtenir una patent genòmica als USA. I precisament ho fa utilitzant arsenal de dades epigenètiques. Després de cinc anys sense beneficis i sense haver apostat per les patents, han trobat un forat i el podrien fer més gran. Cal estar atents.
In
“Round Hill” (1977), the light is a harsh glare, enveloping five
languid bathers in the Caribbean in a self-contained, enclosed moment of
time and place. The figure in the foreground turns away from us, so we
see only the back of his head; the others are self-absorbed, expressions
hidden behind sunglasses.