Real-Time, Multiplexed SHERLOCK for in Vitro Diagnostics
Durant la pandèmia es va produir una innovació notable a la tecnologia de proves diagnòstiques. Va passar desapercebuda per alguns, però no pels que llegiu aquest blog. Es tracta d'utilitzar CRISPR que inicialment s'ha desenvolupat per a edició genètica, per a la detecció d'àcids nucleïcs en un sol pas, i per tant també de Sars-COV-2. La dificultat d'aplicació que tenia aquella prova, que va rebre el vist-i-plau de la FDA era que el procés no es podia automatitzar, i per tant calia amplificació prèvia. Ara acaba de publicar-se la prova definitiva que pot capgirar moltes coses, i que per tant pot canviar la funció de producció dels laboratoris. La troballa d'enzims termoestables ha estat la qüestió clau per a l'èxit de l'equip de Feng Zhang.
Cal dir que la companyia rival, Mammoth Biosciences, de Jennifer Doudna, també té en marxa una prova similar: DETECTR BOOST.
I ara ja ha començat el procés de patentar enzims. Tant per una empresa com per l'altre. I això esdevé inadmisible si el que es pretén patentar la natura o variacions sobre la natura. Però ningú se n'està adonant , als USA ja s'ha fet tard, però a Europa encara hi som a temps.
N'estic segur que les patents, una vegada més, limitaran l'accés a aquesta gran innovació.
Cartier-Bresson
PS. Per si voleu saber com funciona DETECTR BOOST
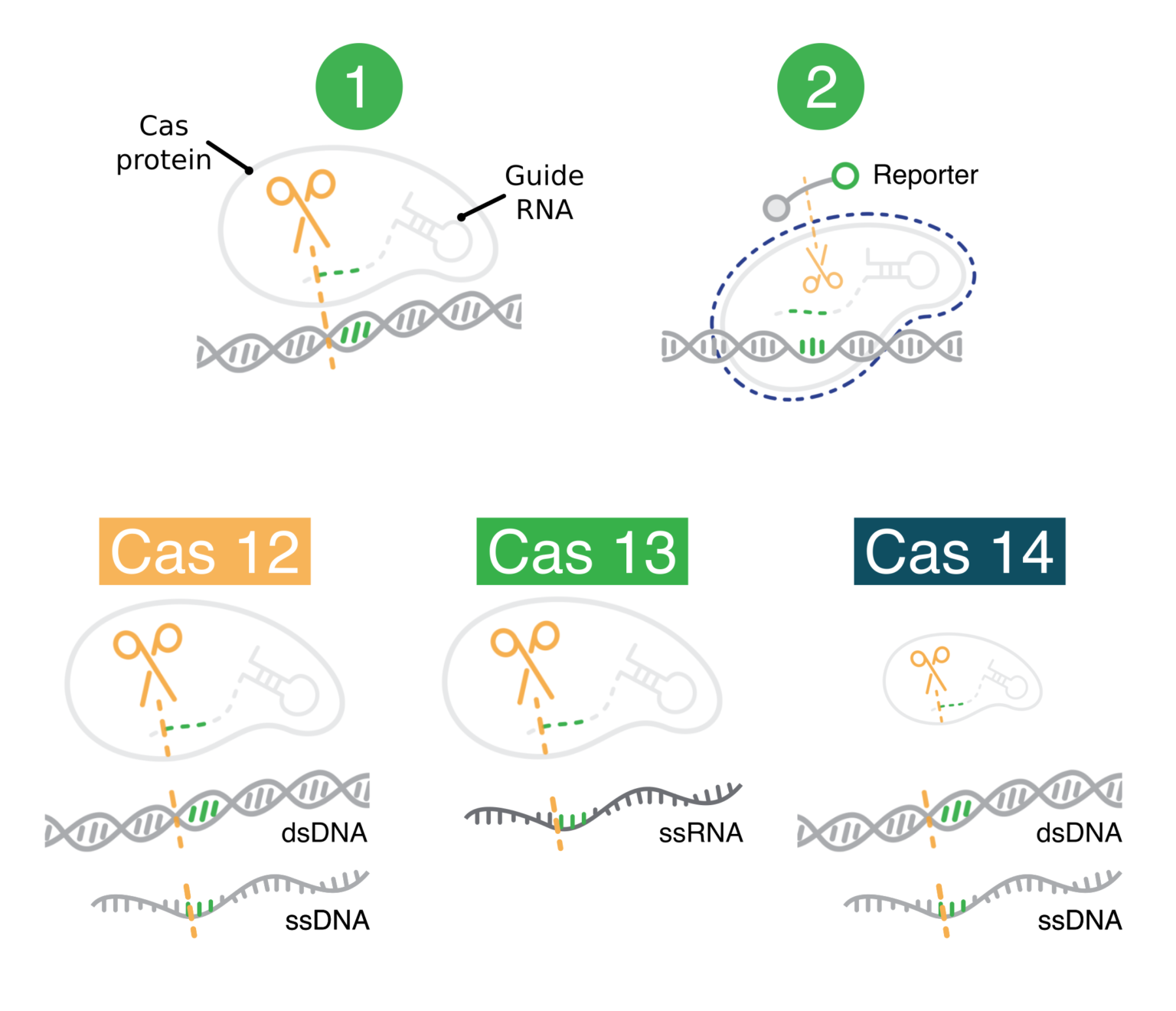
PS. Per tal de comprendre l'abast del que signifiquen les proves diagnòstiques basades en CRISPR el millor és consultar un article de revisió com aquest. I la taula següent conté les dades bàsiques:
Name | Enzyme | Preamplification | Assay timea (min) | Sample preparation | Readout | Applications | LODc (mol l−1) | LODc (copies per ml)3 | References |
|---|
CRISPR type II |
NASBACCb | Cas9 | NASBA | 120–360 (one pot) | Column-based or crude extraction | Colometry | Discrimination between African and American ZIKV | 1.0 × 10–15 | 6.0 × 105 | 25 |
CRISPR–Chip | Cas9 | – | 15 | Column-based | Electrochemical | Detection of gDNA from cell lines and DMD patients | 2.3 × 10–15 | 1.4 × 106 | 45 |
CRISDA | Cas9 nickase | SDA | 90 | Column-based | Fluorescence | Detection of gDNA; breast-cancer-associated SNPs in cell lines | 2.5 × 10–19 | 1.5 × 102 | 27 |
FLASH | Cas9 | PCR | NS | Column-based | NGS | Detection of gDNA; antimicrobial resistance genes in clinical samples | 1.9 × 10–18 | 1.1 × 103 | 71 |
CAS-EXPAR | Cas9 | EXPAR | 60 | Chemical (phase separation) | Fluorescence | Sensing of methylated DNA; L. monocytogenes mRNA | 8.2 × 10–19 | 4.9 × 102 | 91 |
Cas9nAR | Cas9 nickase | Strand-displacing DNA polymerase | 60 | Column-based | Fluorescence | Detection of bacteria (S. typhimurium, E. coli, M. smegmatis, S. erythraea); detection of KRAS SNPs in cell lines | 1.7 × 10–19 | 1.0 × 102 | 111 |
CRISPR type V |
DETECTR | Cas12a | RPA | 10 (RPA) and 60–120 (CRISPR) | Crude extraction | Fluorescence | Detection of HPV16 and HPV18 in human samples | 1.0 × 10–18 | 6.0 × 102 | 36 |
Cas14-DETECTR | Cas14 (Cas12f) | PCR | NS (PCR) and 120 (CRISPR) | Crude extraction | Fluorescence | Detection of HERC2 SNPs in human samples | n.s. | 6.0 × 103 | 41 |
HOLMES | Cas12a | PCR | 88 (PCR) and 15 (CRISPR) | Column-based | Fluorescence | SNP discrimination in cell lines and human samples; detection of viruses (PRV, JEV); virus-strain discrimination | 1.0 × 10–17 | 6.6 × 103 | 37,38 |
CRISPR-materials | Cas12a | RPA | 40 (RPA) and 240 (CRISPR) | Synthetic targets | Fluorescence or μPAD (visual and electronic) | Detection of EBOV synthetic RNA | 1.0 × 10–17 | 6.6 × 103 | 79,80 |
CDetection | Cas12b | RPA | 10 (RPA) and 60–180 (CRISPR) | Synthetic targets or crude extraction | Fluorescence | Detection of HPV16; human ABO blood genotyping; BRCA1 and TP53 SNPs | 1.0 × 10–18 | 6.0 × 102 | 112 |
HOLMESv2 | Cas12b | LAMP | 40 (LAMP) and 35 (CRISPR) or 120 (one pot) | NS | Fluorescence | SNP discrimination in cell lines; RNA virus detection (JEV); human mRNA and circular RNA detection; DNA methylation | 1.0 × 10–17 | 6.0 × 103 | 39 |
E-CRISPR | Cas12a | – | 30–180 | Synthetic targets (nucleic acids) | Electrochemical | Detection of viruses (HPV16, PB19) and protein (TGF-ß1) | 5.0 × 10–11 | 3.0 × 1010 | 77 |
CRISPR type VI |
– | Cas13 | – | NS | NS | Fluorescence | Detection of human mRNA; detection of bacteriophage λ-RNA | 1.0 × 10–12 | 6.0 × 108 | 30,31 |
SHERLOCK | Cas13 | NASBA or RPA | 132 (NASBA) or 120 (RPA) and 60–180 (CRISPR) | Column-based or crude extraction | Fluorescence | Detection of viruses (ZIKV, DENV) and bacteria (E. coli, K. pneumoniae, P. aeruginosa, M. tuberculosis, S. aureus); discrimination between virus strains; detection of SNPs | 2.0 × 10–18 | 1.2 × 103 | 32 |
SHERLOCKv2b | Cas13 | RPA | 60 (RPA) and 60–180 (CRISPR) or 60–180 (one pot) | Column-based or crude extraction | Fluorescence or lateral flow | Detection of viruses (ZIKV, DENV) and bacteria (P. aeruginosa, S. aureus); discrimination between virus strains; detection of SNPs | 8.0 × 10–21 | 4.8 | 22,34 |
SHINEb | Cas13 | RPA | 50 (one pot) | Crude extraction | Fluorescence or lateral flow | Detection of SARS-CoV-2 | 8.3 × 10–18 | 5.0 × 103 | 62 |
STOPCovidb | Cas12b | LAMP | 60 (one pot) | Crude extraction | Fluorescence or lateral flow | Detection of SARS-CoV-2 | 3.3 × 10–18 | 2.0 × 103 | 63 |
CARMEN | Cas13 | PCR or RPA | 20 (RPA) and 180 (CRISPR) | Column-based | Fluorescence | Detection of 169 viruses; subtyping of influenza A strains; detection of HIV drug-resistant mutations | 9 × 10–19 | 5.4 × 102 | 67 |
APC-Cas | Cas13 | Allosteric-probe-initiated amplification with DNA polymerase | 110 (APC) and 30 (CRISPR) | None | Fluorescence | Detection of S. enteritidis | One colony-forming unit | – | 92 |
| | Cas13 | – | <240 | Column-based | Electrochemical | Detection of microRNAs (miR-19b and miR-20a) | 1 × 10–11 | 6.0 × 109 | 46 |
PECL-CRISPR | Cas13 | EXPAR | 30 (CRISPR), 30 (phosphorylation of pre-trigger), 30 (EXPAR) | Column-based | Electrochemiluminescence | Detection of microRNAs (miR-17, let‐7 family miRNAs) | 1.0 × 10–15 | 6.0 × 105 | 78 |
- NS, not specified; APC-Cas, allosteric probe-initiated catalysis and CRISPR-Cas13a system; BRCA1, breast cancer 1 gene; circRNA, circular RNA; Cas9nAR, Cas9 nickase-based amplification reaction; CRISDA, CRISPR–Cas9-triggered nicking endonuclease-mediated strand-displacement amplification; DENV, dengue virus; DMD, Duchenne muscular dystrophy; EBOV, Ebola virus; E. coli, Escherichia coli; HERC2, HECT and RLD domain containing E3 ubiquitin protein ligase 2 gene; HPV, human papillomavirus; JEV, Japanese encephalitis virus; K. pneumoniae, Klebsiella pneumoniae; KRAS, KRAS proto-oncogene GTPase; M. smegmatis, Mycobacterium smegmatis; M. tuberculosis, Mycobacterium tuberculosis; PECL, portable electrochemiluminescence chip; P. aeruginosa, Pseudomonas aeruginosa; PB19, parvovirus B19; PRV, pseudorabies virus; S. erythraea, Saccharopolyspora erythraea; S. aureus, Staphylococcus aureus; S. enteritidis, Salmonella enteritidis; S. typhimurium, Salmonella typhimurium; TP53, tumour protein P53 gene.
- aAssay time indicates the approximate incubation time most frequently used in the referred study (different assay times can be reported, depending on the intended sensitivity and the readout).
- bPOC compatibility indicates whether the entire assay as reported—including sample preparation (that is, crude extraction) and readout—can be performed at POC or in the field with minimal equipment.
- cLimits of detection cannot always be directly compared across studies, in particular because some studies did not report how the LOD was determined, or reported the target concentration either in the transport media of the sample or in the final reaction. The LODs shown here reflect the optimal LODs reported. In general, LODs depend on the type of input material (raw or synthetic), type of readout and incubation time.