Ten Years of Incidental, Secondary, and Actionable Findings
Population genomic screening for three common hereditary conditions : a cost-effectiveness analysis.
El cribratge genètic poblacional s'ha estat realitzant des de fa anys en alguns entorns, molt pocs. Fa cinc anys ja vaig explicar el cas de Geisinger, que ara pertany a Kaiser Permanente. Aquí hi trobareu totes les entrades d'aquest blog sobre proves genètiques.
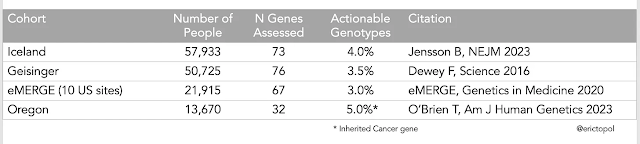
Els resultats en diferents entorns diuen que entre el 3 i el 5% de la població tenen alguns gens amb variants patogèniques sobre els que a data d'avui podem actuar per capgirar el seu impacte. És el que en diuen "actionable". Aquesta és la taula:
Han seqüenciat el genoma o exoma i han informat només d'un nombre determinat de gens, entre 32 i 76. I això ve d'un document de posició que explica quants gens són "actionables" i a data d'avui
són 83. L'origen cal buscar-lo als Genome Wide Association Studies, quan encara hi havia confiança en que trobar un gen que provoca una malaltia permetria afrontar-la i després es va veure que això passava en molts poc casos (ara sabem que paga la pena informar només de 83, els 20.000 ).
Al NEJM ens expliquen un estudi que:
A unique aspect of this study was the reporting of life-span data for persons who carried a pathogenic or likely pathogenic variant as compared with those who did not, in an analysis involving data from 27,546 persons who had died by 2020 and life-span estimates for other persons who were at least 65 years of age. Key findings included an observed shortening in median life span among persons who carried these variants (86 years vs. 87 years for those without). Harboring such a variant in a cancer gene was associated with the largest difference in life span (3 years). By the age of 65 years, 10% of the participants with pathogenic or likely pathogenic variants in cancer genes had died. In contrast, in the group of participants without such a variant, 10% had died by 73 years of age.
Quedem-nos amb la idea, trobar aquesta variant patogènica en un gen suposava una diferència de 10 anys de vida. Al món hi ha dos països que fan cribratge per BRCA2, Israel (població Ashkenazy) i Islàndia. No n'hi ha enlloc més per ara. Ara bé, què costaria fer un cribratge poblacional d'això? Quin seria el cost-efectivitat?
L'Eric Topol ressenya un estudi on per cancer d'ovari i mama, Lynch sindrome i colesterolemia familiar seria cost-efectiu per:
adults younger than 40 years if the genomic test was at a relatively low cost of $250 or for people 50 years old at $166 per test (summary for patients). We are now at that level whereby whole genome sequencing has dropped to the $200 threshold, and targeted sequencing or arrays can be done at substantially lower costs. It doesn’t appear that we can continue to use cost now as the reason to not obtain information that can be lifesaving.
Ens cal molta més informació i anàlisi d'una qüestió tant complexa. Ens trobem davant d'una situació nova que fa molt pocs anys no ens hauríem imaginat. Cal estar atents al cribratge genètic poblacional, i també a què farem quan trobem una variant patogènica "actionable", perquè si no hi ha acció (que vol dir accés a tractament cost-efectiu), llavors, per què cal trobar? Alimentaríem encara més la genòmica recreativa, que a hores d'ara interessa econòmicament moltíssim a alguns.